SBOL: Open Source based exchange for biotech enthusiasts
Especially in the current time we experience how important the exchange between scientists is. Almost every day there are new findings that need to be discussed and debated.
This applies to the situation in the current pandemic as well as to (biotechnological) research in general.
For example, the current state of research makes it possible not only to completely map – or, as it is called in technical jargon, “sequence” – the genome of bacteria and viruses, but also that of humans and animals. It goes even further: genes can not only be sequenced, but also completely synthesized and edited. Most people are probably familiar with the “Big Bang Theory” episode, in which a glowing goldfish is shown. In this illustration this is not scientifically correct, but it is possible: the fish do not glow themselves, but they fluoresce because the gene of a fluorescent jellyfish or sea anemone has been inserted. Thanks to this knowledge, suppliers like PetCo & Co. now have such a “GloFish” in their product range.
For example, in order to be able to exchange such findings with other biotechnology enthusiasts like these in a better and uncomplicated way, a group of scientists has joined forces and developed an international standard: The Synthetic Biology Open Language, abbreviated SBOL. SBOL is subtly divided into two parts:
On the one hand, there is the graphic part “SBOL Visual”: This describes which symbols are used for which functional components of DNA, RNA, etc. – short the “Central Dogma of Molecular Biology” plus X – are used. On the other hand, the data processing part is SBOL Data. This uses already existing principles from information technology such as the automatic conversion of base pair sequences into symbols.
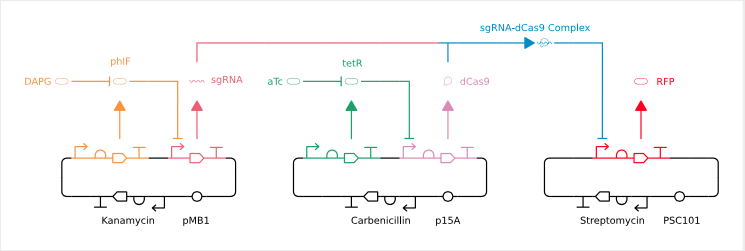
Picture Description: Three DNA rings – also known as plasmids or mini-chromosomes – on which various genes and the interactions between them and their products are shown: A bent arrow (promoter), a semicircle (ribosome binding site), a full arrow (coding part) and a T (terminator) in each plasmid result in a gene. The black areas of the plasmids are responsible for organisational tasks such as self-preservation and reproduction of the plasmids in the living organism.
A red fluorescent protein – RFP – is produced from the gene marked in red. If the protein dCas9(pink) binds to a sgRNA(coral)(blue), the production of RFP is blocked because the connection makes the promoter inaccessible. From the green and orange marked gene proteins (phlF and tetR) are formed which in turn prevent the synthesis of dCas9 and sgRNA. With the addition of DAPG(orange) or aTc(green), these proteins can be manipulated so that they no longer inhibit the production of sgRNA and dCas9, which is ultimately a chemical on-off switch.
But what use is an “open language” if it is not easily accessible. Hardly any biologist, microbiologist or geneticist “speaks” Python, i.e. cannot easily work with the DNAplotlib library. So it had to be something simpler. And something simpler was delivered: Now researchers no longer have to learn a programming language to use the symbols professionally. Because an official font was added to the SBOL package.
We continue to work on SBOL and are constantly adding new symbols and functionalities based on new scientific findings. It is also planned to transfer the symbols to the Unicode table.
If you want to actively participate in development, you can do so in the official GitHub project. More information is also available on the official SBOL page or in this paper.
Der ATIX-Crew besteht aus Leuten, die in unterschiedlichen Bereichen tätig sind: Consulting, Development/Engineering, Support, Vertrieb und Marketing.